-
Notifications
You must be signed in to change notification settings - Fork 3
AK K64G Example Ensemble Simulation
"Paging Dr. Kenny Loggins! You are entering the Danger Zone!"
This advanced demo illustrates how to execute a single-pixel ELM v2 simulation at an NGEE-Arctic study site (i.e. Utqiagvik, Council, Kougarok, Teller). This example illustrates how to configure, run, edit then re-run ELM ensemble simulations. We will start with the Kougarok site but the same approach can be used at any of the four main sites (i.e. Utqiagvik, Council, Kougarok, Teller). The initial demo will demonstrate how to run a shorted ensemble experiment as well as how to setup a proper ensemble simulation that you can run at a later time. As usual, the simulations are controlled by a wrapper script that utilizes the Offline Land Model Testbed, developed and maintained by Dan Ricciuto (ricciutodm@ornl.gov). OLMT controls the creation, configuration, and execution of each ensemble simulation. At the completion of each simulation, the wrapper calls the "ncrcat" command to concatenate all transient simulation years and model variables into a new output file called "ELM_output.nc" within each ensemble member directory. Once complete, the ensemble output will be analyzed in a Jupyter notebook using the provided Jupyter-Lab container environment.
Part 1: Running a short ensemble simulation to demonstrate setting up, running, and viewing ensemble output
First, you will need to download the ELM container designed to run ensembles. You can copy and paste the following code in your terminal window
docker pull fasstsimulation/elm-builds:elm_v2-for-ngee-serial_multiarch
To perform a quick ensemble, you can copy and paste the following code in your terminal window
docker run -t -i --hostname=docker --user modeluser -v elmdata:/inputdata -v elmoutput:/output fasstsimulation/elm-builds:elm_v2-for-ngee-serial_multiarch /scripts/OLMT_ens_docker_example.sh --site_name=kougarok --ad_spinup_years=10 --final_spinup_years=20 --transient_years=5 --num_ens=6 --num_groups=3
This will execute an ensemble with the following options:
case_prefix=OLM_ens (default, can be changed. We will explore changing this below)
site_name=kougarok
ad_spinup_years=10
final_spinup_years=20
transient_years=5
num_ens=6
num_groups=2
This translates to 10 years of accelerated decomposition spinup, 20 years of standard spinup, and a 5 (1850-1855) year transient simulation per ensemble member. The additional options are the number of parameter ensembles (6) and the number of MPI groups (2) to run these cases.
The default file to control parameter options (param_list_example_kougarok) is located in the /scripts folder in the ELM container. We can view this later, for now here is a copy of what that file contains
flnr 11 0.07 0.145
flnr 12 0.07 0.145
vcmaxha 11 65000 75000
vcmaxha 12 65000 75000
jmaxha 11 45000 57000
jmaxha 12 45000 57000
roota_par 11 4.0 10
roota_par 12 6 15
mbbopt 11 1 13
mbbopt 12 1 13
slatop 11 0.009 0.045
slatop 12 0.009 0.045
Once you start the ensemble simulations and after the model builds and the cases start executing, you will see a new "UQ" folder show up in your "elmoutput" docker volume. Inside you will see the three core case groups show up as the execute, AD spinup, spinup and transient

You may also note some warning messages during the model execution that are safe to ignore, e.g.

When the simulation completes you will see a message similar to this show up in your terminal screen

You will also see that the transient directory in your elmoutput/UQ folder will contain 6 ensemble members, as well as a copy of the file that controlled the parameter perturbation experiment used in this short demo

If you have not already done so, create a new docker volume called "elmwork" either in the Docker Desktop app or by typing docker volume create elmwork in your open terminal
To view the output from the initial ensemble output, you need to start the elmlab_3.3.2 container (if its not already running on your host machine)
docker run -p 8888:8888 -v elmdata:/home/jovyan/inputdata -v elmoutput:/home/jovyan/output fasstsimulation/fasst_simulation_tools:elmlab_3.3.2
One the container loads and you open the browser tab to enter into the Jupyter-Lab environment, you can navigate to the output location fo the new ensemble run. Do this by opening the output folder in the directory tree on the left

You can then navigate to the ensemble output by going to /output/cime_run_dirs/UQ/

The transient results are in OLMT_ens_AK-K64G_ICB20TRCNPRDCTCBC, where you will see a series of sub-directories (e.g. g00001, g00002, ...)

You will also see the parameter settings file used for the ensemble simulation.
To plot the results of this first ensemble simulation, navigate back to home by clicking the folder icon in the path browser

Once back, navigate into the scripts folder and open the "plot_ELM_ensembles.ipynb" notebook

Once you open the "plot_ELM_ensembles.ipynb" notebook, you can start analyzing the ensemble simulation results. Note that this notebook has an option to output the figures as a png and pdf file in the work/figures folder. The default behavior is to save the figure files, you can turn this off by changing generate_plots=False, or changing "True" to "False" (with the capital F)

Also note that like the other notebooks, you will select which simulation output you want to review. You do this in step 3 where you select the ensemble output folder you wish to review, e.g.
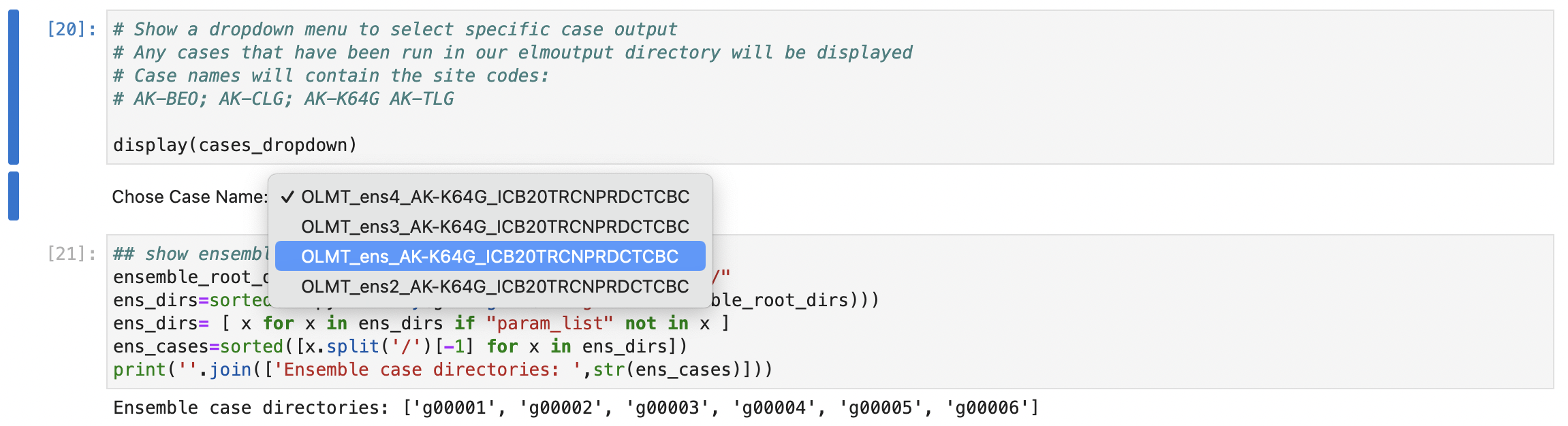
And then in step 4 you will see a list of all ensemble sub-directories printed to the screen

Once you select the output you wish to plot, in step 5 the notebook will load the output from each sub-directory into new objects labeled "output_0, output_1, ...:"
Using these output objects, a series of standardized plots will then generate ensemble plots including all ensemble member output read into Python in step 5, for example a plot of Gross Primary Productivity (GPP)

Using this provided notebook, you can modify the plots, add new variables or change the way the ensemble results are plotted. Just dont forget to save a copy of your new notebook to your host computer before logging out of the Jupyter-Lab environment!
You can revise the previous simulation experiment by increasing the length of the spinup and transient runs. For example, you could modify the spinup and transient run length. The below example will run a 200 year AD and 600 final spinup, and simulate then full transient period (1850-2014) for a similar 6-ensemble simulation
docker run -t -i --hostname=docker --user modeluser -v elmdata:/inputdata -v elmoutput:/output fasstsimulation/elm-builds:elm_v2-for-ngee-serial_multiarch /scripts/OLMT_ens_docker_example.sh --site_name=kougarok --ad_spinup_years=200 --final_spinup_years=600 --num_ens=6 --num_groups=3
You may also want to experiment with changing the parameter options, including what ELM parameters are considered in the perturbation experiment, what PFTs to target for each site, as well as the parameter ranges. To do this, if not already running, start the jupyter-lab container environment. However in this case you want to also include the "elmwork" volume, e.g.
docker run -p 8888:8888 -v elmdata:/home/jovyan/inputdata -v elmoutput:/home/jovyan/output -v elmwork:/home/jovyan/work/ fasstsimulation/fasst_simulation_tools:elmlab_3.3.2
After you have run the default example ensemble case you can copy the file that controls the parameter options from that case directory to the "work" directory in the jupyter-lab environment. To do this navigate, go to the output/cime_run_dirs/UQ/OLMT_ens_AK-K64G_ICB20TRCNPRDCTCBC and copy the file labeled param_list_example_kougarok Go back to the root of the "work" directory and paste the file
Once you copy the param_list_example_kougarok file to "work" you can open the file in the viewer by "double-clicking" the file. It will open in a new tab on the right, e.g.

You may want to right-click and "duplicate" this file, and rename it e.g. param_list_example_kougarok_v2 so that you can keep a copy of the original file as well as your modified files.
You can also add new parameters to the ensemble experiment. For example, you could add in some parameters that control canopy radiation transfer, e.g.

MORE HERE