-
Notifications
You must be signed in to change notification settings - Fork 0
Commit
This commit does not belong to any branch on this repository, and may belong to a fork outside of the repository.
- Loading branch information
1 parent
72fcf3f
commit 7491a42
Showing
28 changed files
with
1,145 additions
and
109 deletions.
There are no files selected for viewing
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| Original file line number | Diff line number | Diff line change |
|---|---|---|
| @@ -1,14 +1,111 @@ | ||
| \brief New module created by make-module.py on Mon Mar 4 17:26:41 2024 | ||
| [](https://pubmed.ncbi.nlm.nih.gov/xyz/) #TODO: Replace xyz with PubMed ID | ||
|
|
||
| Fill in this file with a description of your module. | ||
| [](https://doi.org/10.5281/zenodo.10360718) | ||
|
|
||
| # Info | ||
|
|
||
| _Author(s)_: (full names of author(s)) | ||
| # **NestOR: Nested Sampling-based Optimization of Representation for Integrative Structural Modeling** | ||
|
|
||
| _Maintainer_: (GitHub name of active maintainer) | ||
| 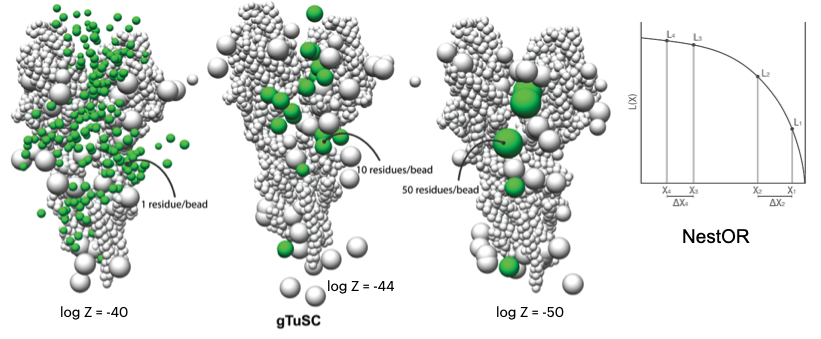 | ||
|
|
||
| _License_: None | ||
| ## **Installation:** | ||
| ### **Dependencies:** | ||
| * IMP (compiled from the source code). See [IMP installation](https://github.com/salilab/imp) | ||
| * Python libraries: `numpy`, `mergedeep`, `mpi4py`, `matplotlib`, `pyyaml` | ||
|
|
||
| _Publications_: | ||
| - None | ||
| ### **NestOR installation:** | ||
| 1. Compile IMP from the souce code to your choice of directory | ||
| 2. Clone this repository and replace `imp/modules/nestor/` with it | ||
| 2. **_(deprecated)_** Replace the `macros.py` in `imp/modules/pmi/pyext/src/` with the `macros.py` in the current repository. Make sure the file is named `macros.py` in the destination directory. Similarly, replace the restraints directory in `imp/modules/pmi/pyext/src/` with the restraints directory in the present repository. | ||
| **_#TODO: Remove deprecated point_** | ||
|
|
||
| ## **Running NestOR:** | ||
|
|
||
| ### Inputs | ||
|
|
||
| (See also `examples/`) | ||
| 1. Split the crosslinks into sampling and evidence calculation subsets using `python pyext/src/xl_datasplitter.py {path}` where, path refers to the path of the target crosslinking file. | ||
| 2. Make the modeling script in the form as shown in the `examples/modeling.py`. One will also need to make separate topology files for different candidate representations. | ||
| _* Make sure that the restraints that are to be used to inform the likelihood have `weight=0`, and these are added to a separate list that is passed to the replica exchange macro as `nestor_restraints` argument_. | ||
| _* Ensure the modeling script looks similar to the one in `example/`. Specifically, ensure that the modeling instructions are enclosed in a function that is called so that the terminal stdout of the modeling is not returned to the terminal. One can use `contextlib` as shown in the example._ | ||
| 4. Set appropriate parameters in the `nestor_params.yaml` file. | ||
|
|
||
| ### Run command | ||
|
|
||
| 1. Run the NestOR wrapper as follows: | ||
| ```python pyext/src/wrapper_v5.py {nestor_param_path} {mode}``` | ||
| where, `nestor_param_path` refers to the absolute path to the `nestor_params.yaml`file and mode refers to the mode of representation (`manual`/`topology`). The default choice is topology (does not need to be mentioned), If the representation is defined in the modeling script, use manual argument. | ||
|
|
||
| 2. Run the following command: | ||
| ```python figure_scripts/plot_evidence_proctime_together.py {path} {name}``` | ||
| where, `path` refers to the parent_dir in NestOR params file and `name` refers to the name of the assembly for which the representations is being optimized. | ||
|
|
||
| **Note** | ||
|
|
||
| _One_ `NestOR run` corresponds to the set of all nested sampling runs for all candidate representations._ | ||
| One can also compare results from `NestOR runs` with different parameter settings by running `python pyext/src/compare_runs_v2_w_pyplot.py {comparison_title} run_set1 run_set2 ...` where comparison_title is the title for the runs to be compared, run_set1 and run_set2 are the NestOR runs to be compared. | ||
|
|
||
| ## Outputs | ||
|
|
||
| ### Plots | ||
|
|
||
| Step 1 in the Run command above, _i.e._ one NestOR run generates these plots: | ||
|
|
||
| 1. **Evidence**: The plot (`*_params_evidence_errorbarplot.png`) shows the mean values of evidence for all the candidate representations along with errorbars showing the standard error on the mean. | ||
| 2. **MCMC per-step time**: The plot (`*_params_persteptime.png`) shows the time required to sample one MCMC step per run. This is computed as `(time taken for iteration 0)/((number of initial frames)*(number of MCMC steps per frame))` | ||
|
|
||
| Step 2 in the `Run command` above generates this additional plot. | ||
|
|
||
| 3. **Evidence and MCMC per-step time per representation** : The plot (`*sterr_evi_and_proctime.png`) compares evidences and their sampling efficiency across representations. | ||
|
|
||
| ### Output YAML file | ||
|
|
||
| This file is generated upon completion of step 1 in the `Run command` above. | ||
|
|
||
| ## Choice of NestOR parameters | ||
|
|
||
| **Evidence related:** | ||
| - log_estimated_evidence: `float` | ||
| _The estimated evidence value represented as natural logarithm of the estimated evidence_ | ||
| - obtained_information: `float` | ||
| _Information obtained from the nested sampling run_ | ||
| - analytical_uncertainty: `float` | ||
| _The analytical uncertainty associated with evidence estimation for a run by nested sampling_ | ||
|
|
||
| **Efficiency related** | ||
| - mcmc_step_time: `float` | ||
| _Time taken per MCMC step. This is computed as `(time taken for iteration 0)/((number of initial frames)*(number of MCMC steps per frame))`_ | ||
| - nestor_process_time: `float` | ||
| _Wall clock time taken by a nested sampling run to finish, represented in seconds_ | ||
|
|
||
| **Termination related** | ||
| - exit_code: `int` (0, 11, 12, 13) | ||
| _Exit code for a nested sampling run_ | ||
| - termination_mode: `str` | ||
| _Cause for run termination_ | ||
| - failed_iter: `int` | ||
| _Number of times Replica Exchange failed to obtain a sample from constrained prior in the current iteration of nested sampling_ | ||
| - last_iter: `int` | ||
| _Iteration count (number of iterations) when nested sampling terminated_ | ||
| - plateau_hits: `int` | ||
| _Number of consecutive times the nested sampling protocol detected a plateau in the estimated evidence_ | ||
|
|
||
| **Exit codes:** | ||
| - Exit code 0: Run terminated normally. | ||
| - Exit code 11: Run terminated due to either a shuffle configuration error or NaN was encountered in the likelihoods. The run will be restarted automatically. | ||
| - Exit code 12: Run terminated as NestOR ran out of maximum allowed iterations. The run will not be restarted. | ||
| - Exit code 13: Run terminated due to *Math domain error* in analytical uncertainty calculation. This happened probably because the run terminated too early resulting in a negative value for H. | ||
|
|
||
| **_If a run terminates with `exit code = 12`, the run is considered incomplete (and is not rerun) and its results are not considered valid, i.e. these are not plotted and not used to infer optimal representation. Results from runs with exit codes 0 and 13 are used to infer the optimal representation_** | ||
|
|
||
|
|
||
| ## **Information** | ||
| **Author(s):** Shreyas Arvindekar, Aditi Pathak, Kartik Majila, Shruthi Viswanath | ||
| **Date**: April 7th, 2023 | ||
| **License:** [CC BY-SA 4.0](https://creativecommons.org/licenses/by-sa/4.0/) | ||
| This work is licensed under the Creative Commons Attribution-ShareAlike 4.0 | ||
| International License. | ||
| **Last known good IMP version:** `not tested` | ||
| **Testable:** Yes | ||
| **Parallelizeable:** Yes | ||
| **Publications:** Arvindekar, S, Pathak., A.S., Majila. K.M., Viswanath, S. Optimizing representations for integrative structural modeling using bayesian model selection. DOI: [10.1093/bioinformatics/btae106](https://doi.org/10.1093/bioinformatics/btae106). | ||
| **_#TODO: Make the publication in appropriate format_** |
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| Original file line number | Diff line number | Diff line change |
|---|---|---|
| @@ -1,3 +1,3 @@ | ||
| required_modules = 'container:core' | ||
| required_dependencies = '' | ||
| optional_dependencies = '' | ||
| required_modules = "container:core:isd:mmcif" | ||
| required_dependencies = "" | ||
| optional_dependencies = "" |
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| Original file line number | Diff line number | Diff line change |
|---|---|---|
| @@ -1,3 +1,3 @@ | ||
| set(pyfiles "") | ||
| set(pyfiles "nude_modeling.py") | ||
| set(cppfiles "") | ||
| set(cudafiles "") |
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| Original file line number | Diff line number | Diff line change |
|---|---|---|
| @@ -1,3 +1,3 @@ | ||
| set(pyfiles "nestor.py") | ||
| set(pyfiles "compare_runs_v2_w_pyplot.py;nestor.py;plot_evidence_proctime_together.py;wrapper_v6.py;xl_datasplitter.py") | ||
| set(cppfiles "") | ||
| set(cudafiles "") |
Oops, something went wrong.