The paper is available on Arxiv: https://arxiv.org/abs/1910.07380
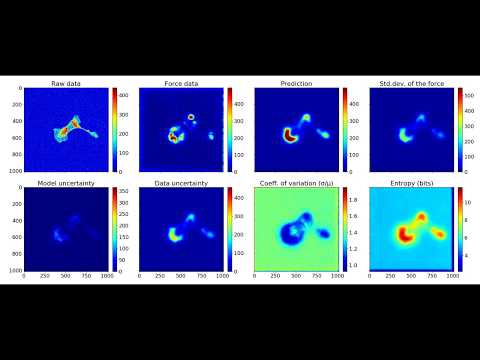
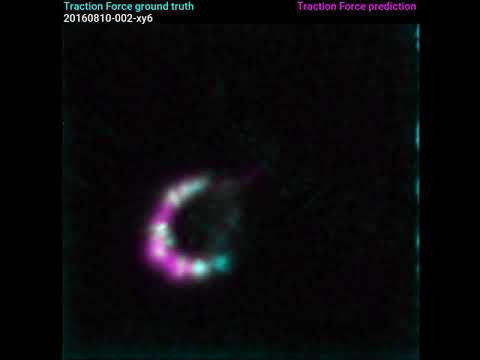
Traction Force Microscopy is a method to determine the tensions a biological cell conveys to the underlying surface. Typically, Traction Force Microscopy requires culturing cells on gels with fluorescent beads, followed by bead displacement calculations. We present a new method allowing to predict those forces from a regular fluorescent image of the cell. Using Deep Learning, we trained a Bayesian Neural Network adapted for pixel regression of the forces and show that it generalises on different cells of the same strain. The predicted forces are computed along with an approximated uncertainty, which shows whether the prediction is trustworthy or not. Using the proposed method could help estimating forces in the absence of non-trivial calculation of bead displacements and can also free one of the fluorescent channels of the microscope.
This repository gives you access to the code necessary to:
- Train a Bayesian Neural Network for Traction Force prediction.
- Test the neural network on other cell datasets.
- Generate the figures present in the published paper.
Video links:
We set up a Colab notebook for you to try predicting the traction forces on any images of your choice. The computation is done on the cloud for free, no need to install anything. Get started in 15 seconds.
You can access the notebook at the following address: https://colab.research.google.com/drive/16eX9UOFn4cQCL6hqcxgiX5QEm8yAhTaR
git clone https://github.com/wahlby-lab/InSilicoTFM.git
pipenv install
jupyter notebookPlease cite the following paper if you are using or contributing to our method:
N. Pielawski, J. Hu, S. Strömblad and C. Wählby, "In Silico Prediction of Cell Traction Forces," 2020 IEEE 17th International Symposium on Biomedical Imaging (ISBI), Iowa City, IA, USA, 2020, pp. 877-881, doi: 10.1109/ISBI45749.2020.9098359.
@INPROCEEDINGS{InSilicoTFM,
author={N. {Pielawski} and J. {Hu} and S. {Strömblad} and C. {Wählby}},
booktitle={2020 IEEE 17th International Symposium on Biomedical Imaging (ISBI)},
title={In Silico Prediction of Cell Traction Forces},
year={2020},
pages={877-881},
doi={10.1109/ISBI45749.2020.9098359}
}
This repository contains the Roboto font designed by Christian Robertson, published under the Apache 2.0 license (https://fonts.google.com/specimen/Robotogo).